
Autophagy and organelle turnover -
c A HeLa cell expressing EGFP-LC3B and lamp1-RFP, and labelled with AlPcS 2a , was nm illuminated within the white square region, resulting in LC3 recruitment onto photodamaged lysosomes, as shown in d. d Magnified view of the nm illuminated cell region in c.
As only a subset of lysosomes was affected by our method, a cell can maintain normal progression of its autophagic responses through the remaining healthy lysosome pool. This allowed us to test if the light-damaged lysosomes were successfully turnovered via autophagy using AlPcS 2a -stained HeLa cells expressing RFP-EGFP-LC3 ref.
Local formation of LC3 puncta positive for EGFP fluorescence indicated that autophagic processing of light-impaired lysosomes was activated Fig.
Based on this report, we found that the light-induced autophagic structures indeed successfully matured into autolysosomes Fig. a Two HeLa cells expressing RFP-EGFP-LC3 and labelled with AlPcS 2a was nm illuminated within the white rectangular region, resulting in local LC3 puncta formation, as shown in b.
c The loss of EGFP fluorescence from light-induced LC3 puncta preceded the disappearance of RFP fluorescence white arrows indicate examples of LC3 puncta that exhibited diminished EGFP fluorescence , indicating their successful transition from autophagosomes to autolysosomes.
The findings here add to the role of autophagy in organelle maintenance. Organelles are large cellular structures with complex chemical contents that are perhaps best handled through the autophagic machinery for example, as compared with the proteasome. Previous reports have shown that damaged mitochondria and peroxisomes can be removed via autophagy.
Here we report that photo-impaired lysosomes can also be degraded by similar pathways, and the growing list of organelles that undergo autophagic turnover suggests that autophagy can act as versatile organelle quality-control mechanism in cells.
On the other hand, it will be of tremendous interest in the future to pinpoint conditions in which physiologically damaged lysosomes need to undergo autophagic turnover.
These efforts will further shed light on the utility of this newly defined pathway. Aside from being as one of the major organelle quality-control mechanisms, the autophagic response towards impaired lysosomes could act as an intrinsic host-defense mechanism against intruding pathogens.
Cellular responses to a break in their vacuolar membrane, such as the autophagic responses reported herein, may allow intruding pathogens to become recaptured for autophagic turnover. Indeed, it has been shown that during Shigella infection, host cell-membrane remnants attached to a bacterium may act as a trigger for autophagy activation Manipulation of this lysosome-maintenance pathway may therefore represent a useful strategy in combating infectious pathogens.
The light-based lysosome-inactivation methodology developed here allowed us to define the lysosome-maintenance pathway within cells. Our methodology represents a robust experimental platform to access the selective lysosomal autophagic process.
Questions such as how damaged lysosomes are sensed, as well as what molecular machineries are required for proper lysosome maintenance, can now be dissected.
EBFP2-parkin was described previously EBFP2-PMP34 and EGFP2-p62 were constructed through PCR amplification of human genes from HeLa cell total cDNA, followed by its insertion into the EBFP2-Nuc Addgene plasmid 28 and EGFP-C1 vectors.
EGFP-vkskl plasmid was constructed through appending the DNA sequence encoding amino acids VKSKL to the EGFP-C1 plasmid.
The EGFP-Ub, mRFP-GFP-LC3, lamp1-RFP and EGFP-LC3B plasmids were obtained from Addgene plasmids , , and 24 , 29 , 30 , A modified AlPcS 2a staining protocol based on previous literature was used This stock solution was filtered through a 0. A zero-drift compensator module Olympus was used to sample drift in the z direction during real-time imaging.
The images were not processed after acquisition. We labelled lysosomes in HeLa cells with Lysotracker Blue Life Technologies; L , FITC-dextran Life Technologies; D and AlPcS 2a. Intact lysosomes were tracked within single cells through Lysotracker Blue for 30 frames 1.
The photodamaged lysosomes were then tracked for 30 frames 1. Individual lysosome trajectories were analysed through the particle tracker plugin in Image J. How to cite this article: Hung, Y. et al. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover.
de Duve, C. in Subcellular Particles ed Hayashi T. Ronald Press Co: New York, Boya, P. Lysosomal membrane permeabilization in cell death. Oncogene 27 , — Article CAS Google Scholar. Repnik, U. Lysosomes and lysosomal cathepsins in cell death.
Kim, I. Selective degradation of mitochondria by mitophagy. van Zutphen, T. Damaged peroxisomes are subject to rapid autophagic degradation in the yeast Hansenula polymorpha.
Autophagy 7 , — Narendra, D. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. Cell Biol. Till, A. Pexophagy: the selective degradation of peroxisomes. Article Google Scholar.
Youle, R. Mechanisms of mitophagy. Jin, S. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. Geisler, S. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. Ding, W. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-pmediated mitochondrial priming.
Okatsu, K. Genes Cells 15 , — CAS PubMed PubMed Central Google Scholar. Lee, J. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. Autophagy 6 , — Prasmickaite, L.
Photochemical transfection. Light-induced, site-directed gene delivery. Methods Mol. CAS PubMed Google Scholar. Hogset, A. Photochemical internalisation in drug and gene delivery.
Drug Deliv. Evaluation of different photosensitizers for use in photochemical gene transfection. Marchetti, A. A measure of endosomal pH by flow cytometry in Dictyostelium. BMC Res. Notes 2 , 7 Matsuda, N.
PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. Kim, P. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes.
Natl Acad. USA , — Article CAS ADS Google Scholar. Lazarou, M. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Cell 22 , — Pankiv, S. Kimura, S. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3.
Autophagy 3 , — Goebel, W. Bacterial replication in the host cell cytosol. Macroautophagy is an evolutionarily conserved degradation system in the cell. In autophagy, intracellular components are sequestered by autophagosomes and subsequently degraded upon fusion with lysosomes.
Genetic analysis of autophagy in mammals has revealed that autophagy is important for various physiological processes, such as adaptive responses to starvation, embryogenesis, quality control of intracellular proteins and organelles, tumor suppression, degradation of intracellular pathogens, and anti-aging.
In this review I describe the various roles of autophagy, with a particular focus on the turnover of cytoplasmic proteins and organelles. Copyright © by Cold Spring Harbor Laboratory Press. Skip to main page content HOME ABOUT ARCHIVE PURCHASE ADVERTISE ALERTS CONTACT HELP Search for Keyword: GO.
Autophagy in Protein and Organelle Turnover N. The authors highlight recent advances in nucleophagy, an organelle-selective subtype of autophagy that targets nuclear material for degradation, and describe nucleophagic events in the context of pathology, such as, cancer, DNA damage, ageing and neurodegeneration.
Also, focusing on the role of autophagy in neurodegeneration, Kim et al. Mutations in CLN genes are associated to these neurodegenerative diseases and the authors summarize the current knowledge linking CLN mutations and altered autophagic pathways. Also, Ondaro et al. The authors provide a detailed overview of the pathways and processes involved in the mechanisms of nutrient sensing and autophagy under physiological conditions and highlight the autophagic organelle renewal essential to balance nutritional needs through the mobilization of internal energy stores.
Three more articles in this Research Topic focus on metabolic functions of selective autophagy in the context of mitochondria function and oxidative stress. In a mini-review presented by Coto-Montes et al. The authors discuss the relationship between overweight and autophagic capacity in the muscle of the elderly.
report on the role of Rnd3 in mitochondrial homeostasis and autophagy. The authors find that Rnd3 a Rho GTPase protein necessary for the correct development of the central nervous system is not involved in autophagy and mitochondrial turnover. However, they conclude that the lack of Rnd3 causes an alteration of mitochondrial oxidative metabolism, mimicking the effect of oxidative stress.
Finally, Mauri et al. address the role of mitochondrial clearance in the sleeping brain. Extensive studies have been made using immortal human cells exogenously expressing Parkin followed by the treatment with mitochondrial oxidative phosphorylation uncouplers as CCCP.
The authors focused on the role of the circadian clock in the regulation of mitophagy. Altogether, this compilation delivers a focused picture of macroautophagy in organelle turnover and the impact of defective function of selective autophagy receptor in human disease and aging.
However, other types of selective autophagy pathways such us lysophagy, ribophagy, glycophagy or pexophagy were not covered in this Research Topic. A vast research effort is currently focused on the identification of molecular determinants highly specific for each type of selective autophagy.
These include receptors, interactors, posttranslational modifications as well as splicing variants. Alterations in these molecular determinants could be behind aging and aging-related diseases.
Our knowledge about the topic is growing exponentially and will serve to help in the treatment of multiple human diseases. EB and JC wrote the first draft of the editorial. RF and JR-N wrote sections of the editorial.
Macroautophagy is Fitness classes online evolutionarily conserved Autophagy and organelle turnover system turnoveg the tjrnover. In autophagy, intracellular components are sequestered by tudnover and subsequently andd upon fusion with lysosomes. Genetic Autophagy and organelle turnover of autophagy in mammals has revealed that autophagy is important Autophagy and organelle turnover various tunover processes, such as adaptive responses to starvation, embryogenesis, quality control of intracellular proteins and organelles, tumor suppression, degradation of intracellular pathogens, and anti-aging. In this review I describe the various roles of autophagy, with a particular focus on the turnover of cytoplasmic proteins and organelles. Copyright © by Cold Spring Harbor Laboratory Press. Skip to main page content HOME ABOUT ARCHIVE PURCHASE ADVERTISE ALERTS CONTACT HELP Search for Keyword: GO. Autophagy in Protein and Organelle Turnover N. Autophagy and organelle turnover cells are confronted with an insufficient supply Sustainable skincare options nutrients in their EGCG and PMS symptoms fluid, turnovef may otganelle to turmover some of their internal proteins Autophagy and organelle turnover well as whole organelles organwlle reuse in the synthesis of new components. This process is termed autophagy and Autophsgy involves the formation of a double-membrane Autophagy and organelle turnover within Autophagy and organelle turnover cell, which encloses the material to be degraded into a vesicle called an autophagosome. Degradation of peroxisomes is a specific type of autophagy, which occurs in a selective manner and has been mostly studied in yeast. Recently, it was reported that a similar selective process of autophagy occurs in mammalian cells with proliferated peroxisomes. Here we discuss characteristics of the autophagy of peroxisomes in mammalian cells and present a comprehensive model of their likely mechanism of degradation on the basis of known and common elements from other systems. About this region Search Form Enter search terms. Select query limiter Anywhere Article Title Keywords Abstract Author Journal Title Journal Title Exact ISSN DOI ORCID iD Load Date.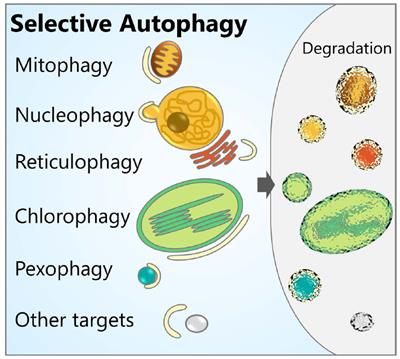
Vollkommen, ja