Autophagy and proteasomal degradation -
Immunoprecipitation was performed by adding 30 μL of GFP-Trap coupled to agarose beads ChromoTek and samples were incubated for 2 h at 4°C with continual rotation.
Beads were subsequently washed five times with Tris-buffered saline containing 0. Proteins were extracted in mM Tris pH 7.
This was followed by incubation with horseradish peroxidase-conjugated secondary antibodies diluted , in PBS containing 0. The immunoreaction was developed using an ECL Prime Kit GE Healthcare and detected with a LAS Luminescent Image Analyzer Fujifilm. Total RNA was isolated using the RNeasy Plant Mini Kit Qiagen , and on-column DNA digestion was performed with DNase I Qiagen.
First-strand cDNA was synthesized from 1 μg of total RNA using the Maxima First Strand cDNA Synthesis Kit Thermo Fisher Scientific. Normalization was done using PP2A AT1G Proteasome activity was determined as described previously Üstün et al.
The dual luciferase reporter assay was performed according to the manufacturer's instructions Dual-Luciferase Reporter Assay System; Promega with slight modifications. Briefly, four leaf discs were homogenized in μL lysis buffer and cleared by centrifugation.
For detection and measurement of the Firefly luciferase activity, 40 μL of the luciferase assay reagent was added to 5 μL of plant extracts. To measure Renilla luciferase activity, 40 μL of the Stop and Glo reagent was added to the mixture. The measurement was performed in a plate reader.
For quantification of water-soaking in HopM1-expressing N. benthamiana leaves, electrolyte leakage assays were done essentially as described Üstün et al.
In brief, four leaf discs 0. Conductivity of the bath solution was measured with a conductometer EcoScan Hand-held Series; Eutech Instruments , and values were expressed as the percentage of maximum values obtained by subsequent boiling of the samples for 30 min.
Data are presented as mean ± sd. The number of biological replicates n is given in the figure legends. Supplemental Figure 1.
Pst -mediated inhibition of proteasome function is dependent on a functional autophagy pathway and independent of the SA pathway. Supplemental Figure 2. TOR inhibitor AZD induces autophagy. Supplemental Figure 3. AZD promotion of bacterial growth is dependent on a functional autophagy pathway.
Supplemental Figure 4. Kinetics of GFP-ATG8a accumulation upon challenge with Pst and bacterial PAMPs. Supplemental Figure 5. Protein expression analysis of T3Es used in the luciferase-based autophagy assay in N. Supplemental Figure 6. Supplemental Figure 7. Transcriptional induction of proteasome subunit genes in response to Pst infection.
Supplemental Figure 8. Proteasome subunit levels do not change in response to PstΔhrcC. Supplemental Figure 9. PAG1-GFP localization upon chemical proteasome inhibition and PstΔhrcC infection. Supplemental Figure Colocalization of PAG1-GFP with RFP-ATG8e and RFP-ATG8g in N.
Proteasome activity and bacterial growth in rpn10 mutant. PAG1-GFP aggregates and punctuate structures are induced upon delivery of HopM1 by Pst D28E. Coimmunoprecipitation of GFP-ATG8a and NBR1 upon Pst infection. Protein abundance of HopM1 upon coexpression with wild-type NBR1 or UBA2 and LIR mutant variants.
Hyperaccumulation of ubiquitinated proteins during Pst infection in nbr Supplemental Table 1. Supplemental References. We thank Sheng-Yang He Michigan State University, East Lansing, MI for the DEX:HopM1-GFP construct and transgenic Arabidopsis lines, Alan Collmer Cornell University, Ithaca, NY for providing the P.
syringae strains used in this study, and Hongyong Fu Academia Sinica, Taiwan for the anti-RPN10 antibody. We also thank Kristiina Mäkinen University of Helsinki, Finland for providing pRDS:FLUC and pRDS:RLUC vectors.
This research was funded by grants from the Swedish University of Agricultural Sciences , the Knut and Alice Wallenberg Foundation , the Carl Tryggers Foundation , and the Swedish Research Council VR to D.
and A. and R. were supported by a grant from the U. Department of Energy Office of Science, Office of Basic Energy Science, Chemical Sciences, Geosciences and Biosciences Division DE-FGER and D. designed experiments. performed experiments. analyzed the data.
contributed novel experimental material. wrote the article. Bibo-Verdugo , B. Targeting proteasomes in infectious organisms to combat disease.
FEBS J. Google Scholar. Book , A. Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes.
Boya , P. Emerging regulation and functions of autophagy. Cell Biol. Chung , T. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci.
Plant J. Cohen-Kaplan , V. p and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. USA : E — E Coll , N. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. Cunnac , S. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae.
USA : — Curtis , M. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. Dagdas , Y. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor.
eLife 5 : e DebRoy , S. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants.
Derrien , B. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Dong , X. Autophagy and viruses: adversaries or allies?
Innate Immun. Eskelin , K. Potyviral VPg enhances viral RNA Translation and inhibits reporter mRNA translation in planta. Gimenez-Ibanez , S. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis.
PLoS Biol. Gladman , N. The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis.
Plant Cell 28 : — Gomes , L. Autophagy in antimicrobial immunity. Cell 54 : — Grefen , C. A ubiquitin promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies.
Guo , Y. AP1 is essential for generation of autophagosomes from the trans-Golgi network. Cell Sci. Hafrén , A. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles.
Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Han , S. Cytoplastic glyceraldehydephosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana.
Plant Cell 27 : — Haxim , Y. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 6 : e Hofius , D. Autophagy as an emerging arena for plant-pathogen interactions. Plant Biol.
Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell : — Ishiga , Y. Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions.
Plant Methods 7 : Jelenska , J. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Kalinowska , K. All roads lead to the vacuole-autophagic transport as part of the endomembrane trafficking network in plants.
Klionsky , D. The mechanism and physiological function of macroautophagy. Kurepa , J. Kwon , S. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Lai , Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens.
Lenz , H. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Levine , B. Autophagy in immunity and inflammation. Nature : — Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants.
Lin , Y. The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN Plant Cell 23 : — Liu , Y. TOR is a negative regulator of autophagy in Arabidopsis thaliana.
PLoS One 5 : e Autophagy regulates programmed cell death during the plant innate immune response. Macho , A.
Subversion of plant cellular functions by bacterial type-III effectors: beyond suppression of immunity. New Phytol. Marshall , R. Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone.
Cell Reports 16 : — Cell 58 : — Mattera , R. AP-4 mediates export of ATG9A from the trans -Golgi network to promote autophagosome formation. Minina , E. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness.
Autophagy and metacaspase determine the mode of cell death in plants. Misas-Villamil , J. Subunit-selective proteasome activity profiling uncovers uncoupled proteasome subunit activities during bacterial infections. Mizushima , N.
Autophagy: renovation of cells and tissues. Mostowy , S. Autophagy and bacterial clearance: a not so clear picture. Munch , D. Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis.
Autophagy 10 : — Nakagawa , T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Noda , T. Autophagy in the context of the cellular membrane-trafficking system: the enigma of Atg9 vesicles. Nomura , K. A bacterial virulence protein suppresses host innate immunity to cause plant disease.
Science : — Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Paul , P. Autophagy and mammalian viruses: roles in immune response, viral replication, and beyond.
Virus Res. Popa , C. The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Plant Sci. Reggiori , F.
Autophagosome maturation and fusion. Shpilka , T. Essential role for the mammalian ATG8 isoform LC3C in xenophagy. Cell 48 : — Smalle , J. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN Plant Cell 14 : 17 — The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling.
Plant Cell 15 : — Svenning , S. Autophagy 7 : — Teh , O. Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. Thompson , A. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Üstün , S. The Xanthomonas campestris type III effector XopJ proteolytically degrades proteasome subunit RPT6.
Autophagy as a mediator of life and death in plants. By this way, recycling of proteins result in the generation of amino acids that are ultimately reused by cells in the synthesis of new proteins. The 26S proteasome contains an additional 19S cap structure that further regulates the internalization of ubiquitylated substrates Lander et al.
The central part of the 19S cap consists of six AAA ATPases Rpt1—Rpt6 forming the Rpt ring that is responsible for substrate binding and unfolding as well as substrate transfer through the channel Collins and Goldberg, Non-ATPase proteins such as Rpn10 and Rpn13 in the 19S cap, possess ubiquitin-binding domains and therefore function as receptors for ubiquitin-labeled substrates Finley, Recent studies showed that ubiquitylation is a reversible phenomenon.
Deubiquitinating enzymes DUBs are proteases that remove ubiquitin or ubiquitin-like molecules from substrates and disassemble polyubiquitin chains. DUBs regulate UPS-mediated degradation in different cellular contexts Reyes-Turcu et al.
Moreover, they play an important role in the control of available free ubiquitin pool in cells, allowing recycling and reuse of ubiquitin. Some DUBs are also responsible for processing newly synthesized ubiquitin precursors Komander et al.
There are three major types of autophagy: Macroautophagy, microautophagy and chaperon-mediated autophagy CMA. In this review, we chose to focus on macroautophagy herein autophagy. CMA and microautophagy were discussed in elsewhere Kaushik and Cuervo, ; Oku and Sakai, Autophagy is characterized by the engulfment of cargo molecules by double-membrane vesicles, called autophagosomes Klionsky, ; Mizushima, , ; Lamb et al.
Following closure, autophagosomes are transported by the microtubule system, leading to their fusion with late endosomes and lysosomes, forming autolysosomes. In this new compartment, sequestered cargos are degraded by the action of lysosomal hydrolases. Building blocks that are generated by hydrolysis of macromolecules e.
Active at a basal level, autophagy is upregulated following a number of stimuli and stress conditions. Amino acid deprivation, serum starvation and growth factor deprivation, hypoxia, exposure to various chemicals and toxins might be counted among stress conditions activating autophagy.
FIGURE 2. Stages of the autophagy pathway for detail, see the text. A Upstream signaling, B membrane nucleation stage, C elongation and closure stage, D autophagosome-lysosome fusion stage.
Most autophagy inducing signals converge at the level of mTOR protein complexes mTORC1 and mTORC2 that coordinate anabolic and catabolic processes Sabatini, ; Saxton and Sabatini, Figure 2. Cellular energy sensor AMPK directly regulates mTOR and therefore contributes to the regulation of the autophagic activity.
mTORC1-dependent phosphorylation of ULK1 and Atg13 Hosokawa et al. A class III phosphatidylinositol 3-kinase PI3K complex, including the lipid kinase VPS34 and the regulatory protein Beclin1, controls the membrane nucleation stage and initial phagophore formation.
Phosphatidylinositol 3-phosphate PtdIns3P that is generated by PI3K activity serves as a landing pad for autophagy-related proteins containing PI3P-binding domains e. Among them WIPI and DFCP1 were involved in the formation of a membrane structure called omegasome or cradle, a structure that creates a platform for the elongation of autophagosome precursor isolation membranes Mauthe et al.
Elongation of the isolation membrane depends on two ubiquitin-like conjugation systems. In the first system, autophagy-related gene 12 ATG12 is covalently conjugated to the ATG5 protein through the action of ATG7 E1-like and ATG10 E2-like proteins.
Then, recruitment of the ATG16L1 protein to ATG dimer results in the formation of a larger complex. Then forming ATGL1 oligomers serve as E3 ligases that conjugate lipid molecules such as phosphatidylethanolamine to ATG8 orthologs MAP1LC3, GATE16, GABARAP Mizushima et al.
Lipid-conjugated ATG8 proteins are required for the elongation, expansion and closure of autophagosome membranes Nakatogawa et al. In order to acquire lytic capacity, autophagosomes fuse with late endosomes or lysosomes.
In mammalian cells, fusion requires lysosomal integral membrane protein LAMP-2, several SNARE proteins e. Following fusion of the outer membrane of autophagosomes, materials contained in the inner membrane are degraded by the action of lysosomal hydrolases Tanida et al.
Building blocks e. are then transported back to cytosol for reuse in the metabolic processes of the cells. Autophagic vesicles engulf targets such as portions of cytoplasm and various cytoplasmic components in a non-selective manner. On the other hand, several selective forms of autophagy have been described Kraft et al.
In most cases, ubiquitylation of the cargo constitutes a key step in the chain of events leading to its autophagic removal Kirkin et al. Selective targets include mitochondria Okamoto et al. By this way, cells control number of the organelles, eliminate dysfunctional components and get rid of potentially harmful aggregates and invaders.
Selectivity is ensured by target-specific autophagy receptors that form a bridge between the ubiquitylated cargo and LC3 component of autophagic membranes. Selective autophagy relies on the recognition and binding capacity of autophagy receptors to various types of cargo, including mitochondria OPTN, NDP52, Tax1BP1, NIX, FUNDC1 Novak et al.
LC3-interacting region LIR is the common motif which allows autophagy receptors to bind lipidated LC3. On the other hand, ubiquitin-associated domain UBA domain on autophagy receptors are responsible for the recognition of ubiquitin decorated cargos Khaminets et al.
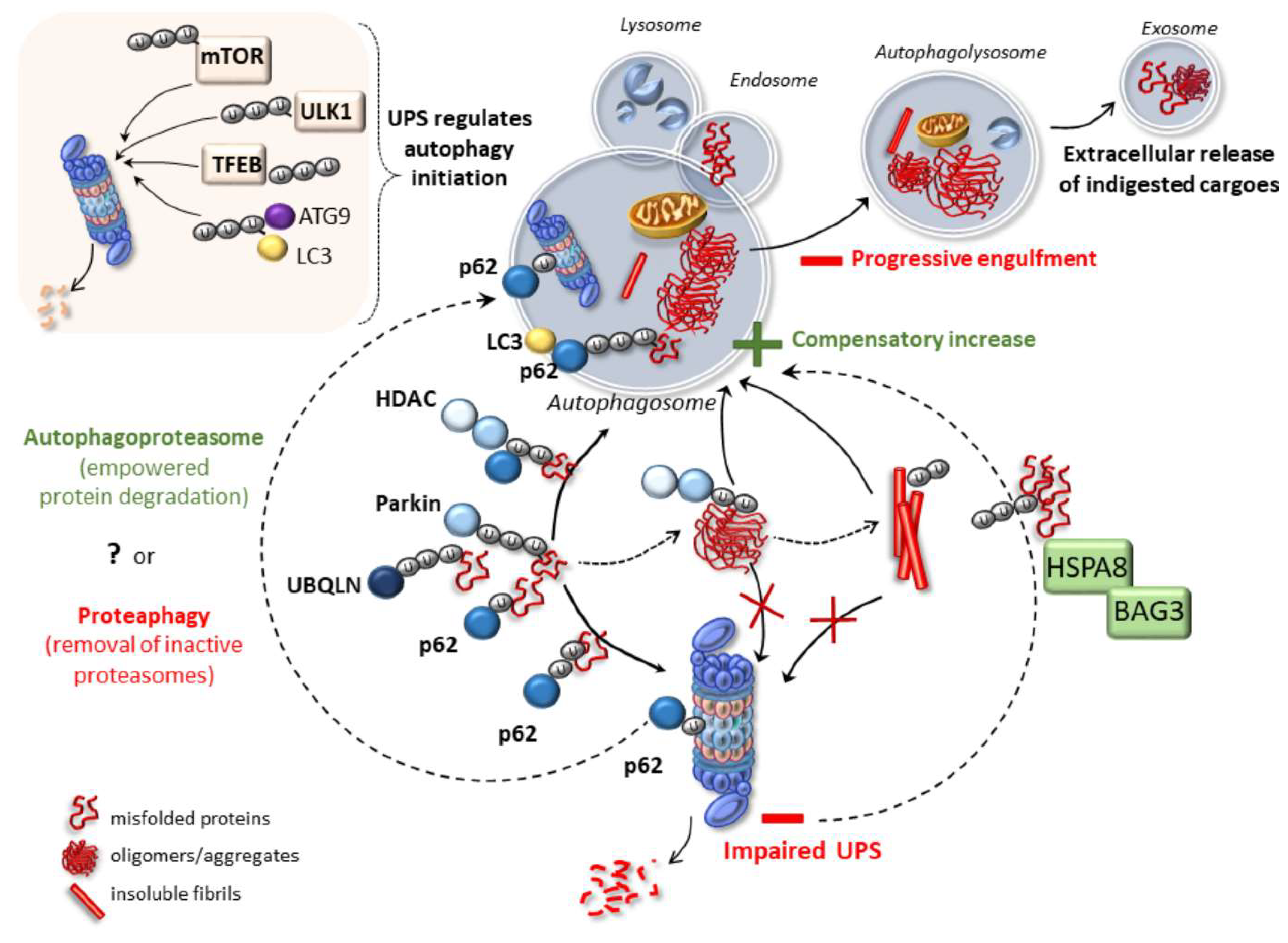
Cargos that are wrapped and packed in autophagosomes are then ready for delivery and degradation in lysosomes. The UPS and autophagy are the two major and evolutionarily conserved degradation and recycling systems in eukaryotes. Although their activities are not interdependent, recent studies show that connections and crosstalks exist between the two systems.
Mitophagy constitutes a prominent example connecting these two degradative systems, yet several other examples exist. In this section, we will summarize biological events involving autophagy and the UPS, and discuss molecular details of the crosstalk mechanisms.
Initial observations about functional connections between the UPS and autophagy systems revealed that inhibition of one led to a compensatory upregulation of the other system. In order to maintain homeostasis, cellular materials that accumulate following inhibition of one degradative system needs to be cleared, at least in part, by the other system Figure 3.
Here, we will give examples of scenarios where these compensation mechanisms are operational. FIGURE 3. The compensatory balance between the activities of autophagy and the UPS in order to maintain cellular homeostasis.
Inhibition of the UPS using various compounds e. Wu et al. For example, inhibition of proteasomal activity by the proteasome inhibitor and chemotherapy agent bortezomib led to an increase in the expression of autophagy genes ATG5 and ATG7, and induced autophagy.
In fact, autophagy gene upregulation depended on an ER stress-dependent pathway that involved eukaryotic translation initiation factor-2 alpha eIF2α phosphorylation Zhu et al.
In another study, proteasome inhibition was associated with an increase in p62 and GABARAPL1 levels by Nrf1-dependent and -independent pathways prior to autophagy activation Sha et al. Autophagy induction following proteasome inhibition correlated with AMPK activation as well.
A number of studies provided evidence that proteasomal inhibition is sensed by both AMPK and mTORC1, two major regulators of autophagy. For instance, in macrophages, epitelial and endothelial cells, proteasome inhibition using chemicals resulted in the activation of AMPK Xu et al.
In some other cancer cell types, CaMKKβ and glycogen synthase kinase-3β GSK-3β were identified as upstream regulators of AMPK activation, proteasome inhibition was linked to a decrease in GSK-3β activity and to the activation of AMPK and autophagy Sun et al.
On the other hand, Torin or rapamycin-mediated inhibition of mTOR stimulated long-lived protein degradation through activation of both UPS and autophagy Zhao et al. In retinal pigment epithelial cells, inhibition of proteasome by lactacystin and epoxomicin was shown to block the AKT-mTOR pathway and induce autophagy Tang et al.
SiRNA-mediated knockdown of Psmb7 gene coding for the proteasome β2 subunit, resulted in enhanced autophagic activity, and it was linked the mTOR activation status of cultured cardiomyocytes Kyrychenko et al. Similarly, impairment of autophagy correlated with the activation of the UPS.
In colon cancer cells, chemical inhibition of autophagy and small RNA mediated knock down of ATG genes resulted in the upregulation of proteasomal subunit levels, including the catalytic proteasome β5 subunit, PSMB5 and led to increased UPS activity Wang et al.
In another study, 3-MA-mediated autophagy inhibition in cultured neonatal rat ventricular myocytes NRVMs increased chymotrypsin-like activity of proteasomes Tannous et al. Since proteasomes were identified as autophagic degradation targets proteaphagy , enhanced proteasome peptidase activity following autophagy inhibition might be associated with the accumulation of proteasomes Cuervo et al.
Yet in several cases, autophagy inhibition correlated with the accumulation of ubiquitylated proteins. For instance in independent studies with ATG5 or ATG7 knockout mice, accumulation of ubiquitylated conjugates were observed, especially in the brain and the liver of the animals Komatsu et al.
Similar results were observed in other animal models such as Drosophila Nezis et al. In line with these data, inhibition of autophagy through siRNA-mediated knockdown of ATG7 and ATG12 in HeLa cells resulted in the impairment of UPS, accumulation of ubiquitylated proteins as well as other important UPS substrates, including p53 and β-catenine Korolchuk et al.
In above-cited papers, autophagy impairment followed by the autophagy receptor p62 accumulation in cells, and played a key role in the observed UPS defects.
Ubiquitylation was proposed to be a common component that directs substrates to the proper degradation system and even contribute to the UPS-autophagy crosstalk Korolchuk et al.
According to this view, proteins that are predominantly linked to Kbased ubiquitin chains are generally directed for degradation through UPS. Conversely, aggregates that are linked to Kbased ubiquitin chains are directed for autophagic degradation. P62 binding capacity was introduced as the critical step in the choice between the UPS and autophagy.
Although, p62 is able to attach both K and Klinked ubiquitin chains through its UBA domain, binding affinity of the protein for Klinked chains seems to be higher Long et al. Due to this dual ubiquitin binding ability, p62 might show UPS inhibitory effects in some contexts. In summary, in the case of a defect in one of the two degradation systems, the other system is upregulated in order to eliminate ubiquitylated protein substrates.
Yet, compensation does not always work and its success largely depends on cell types, cellular and environmental conditions and target protein load.
Function of proteins depend on their proper folding and 3D structures. Various insults, including heat shock, organellar stress, oxidative stress etc. Moreover several disease-related mutations were associated with folding problems.
Failure to refold result in dysfunctional or malfunctional, hence toxic protein accumulations, activation of stress and even cell death pathways. In order to control toxic protein accumulations, an active process of protein aggregate formation comes into play.
Additionally some proteins, including mutant proteins are already prone to form aggregates. Selective clearance of most cytosolic proteins require ubiquitylation. Depending on their solubility, ubiquitylated proteins and protein aggregates are then cleared by the UPS or autophagy.
Soluble fractions of proteins with a folding problem are recognized by the chaperone machinery and directed to the UPS for degradation.
BAG family proteins, especially BAG1, interact with the Hsp70 complex and induce proteasomal degradation of client proteins. On the other hand, clearance of insoluble aggregate-prone proteins require formation of aggresomes.
Ubiquitylation by a number of different E3 ligases, including CHIP, Parkin, HRD1 and TRIM50 prime aggregate-prone proteins Olzmann et al. HDAC6 is another protein that plays a key role in the process of aggresome formation. HDAC6 was shown to provide the link between Kbased ubiquitylated aggregates and microtubule motor protein dynein Matthias et al.
Then, dynein-mediated mechanism direct the aggregates toward microtubule organizing centers MTOCs , resulting in their piling of as aggresomes Johnston et al. Following aggresome formation, direct interaction of adaptor proteins p62 and NBR1 with ubiquitylated aggregates result in their delivery to autophagosomes Ichimura et al.
Another autophagy-related protein, ALFY, was also identified as a player in the selective autophagy and degradation of aggresomes Clausen et al. FIGURE 4. Misfolded proteins can be eliminated by both the UPS and autophagy system.
Misfolded proteins are ubiquitylated and based on the differences in ubiquitin linkages and ubiquitin binding proteins, they are directed for proteasomal degradation or further accumulated in aggresomes.
Aggresomes are selectively cleared by autophagy. An alternative pathway for aggresome formation require Hsp70 partner proteins BAG3 and CHIP Zhang and Qian, Similar to HDAC6, BAG3 binds to dynein, and this directs Hsp70 substrates to aggresomes.
However, BAG3-dependent aggresome formation was not dependent on the ubiquitylation of substrates as in the case of HDAC6, and CHIP E3 ligase activity was dispensible Gamerdinger et al. Yet, E3 ligases such as CHIP were required for BAG3-dependent aggresome clearance by autophagy Klimek et al.
Until so far, we focused on the UPS and autophagy as complementary but independent mechanisms. However, there are cases where components of one system were reported to be a proteolytic target of the other system.
For example, a number of autophagy proteins were regulated through degradation by the UPS. On the other hand, even the whole proteasomes were shown be selective targets of autophagic degradation.
Here, we will give examples of how mutual regulation through proteolysis contributes to the crosstalk and the interplay between the two systems.
Early studies indicated that proteasomes could be degraded in lysosomes Cuervo et al. Later on, plant studies revealed that lysosomal degradation of 26S proteasomes occurred by a specific form of selective autophagy, proteaphagy Marshall et al. RPN10 protein was introduced as an ATG8 interacting plant proteaphagy receptor.
Instead, Cue5 protein in the yeast and its human ortholog TOLLIP, were introduced as selective receptors regulating proteasome clearance by autophagy Lu et al. Moreover, p62 was also described as another proteaphagy receptor Cohen-kaplan et al.
For example, in mammals, amino acid starvation significantly upregulated ubiquitylation of 19S proteasome cap components RPN1, RPN10, RPN13, and led to their pmediated recruitment to autophagosomes Cohen-kaplan et al. Interestingly during carbon or nitrogen starvation, plant and yeast proteasomes were shown to localize in proteasomal storage granules PSGs , protecting them from autophagic degradation during stress Peters et al.
Whether similar mechanisms exist in the mammals is currently an open question. These observations underline the importance of selective degradation of proteasome by autophagy in the control of proteasome numbers as well as overall UPS and lytic activity in cells.
FIGURE 5. Schematic representation of the selective degradation of proteasomes by autophagy. Upon starvation and functional defects proteasomes become ubiquitylated and degraded by autophagic machinery.
Modulation of the half-life of some proteins in the autophagy pathway by the UPS serves as a means to control cellular autophagic activity. For instance, LC3 protein was shown to be processed in a stepwise manner by the 20S proteasome, a process that was inhibited by p62 binding Gao et al.
On the other hand, E3 ligase NEDD4-mediated Klinked ubiquitylation of Beclin1 prevented its binding to the lipid kinase VPS34, and led to its degradation Platta et al. Another E3 ligase, RNF ubiquitylated Beclin1 adding Klinked ubiquitin chains on the protein Xu et al.
Beclin1 ubiquitylation resulted in autophagy blockage in both cases. Conversely, reversal of Beclin1 ubiquitylation by the DUB protein USP19 stabilized the protein under starvation conditions and promoted autophagy Jin et al.
USP10 and USP13 as well as USP9X were characterized as other DUBs that regulated autophagy through control of Beclin1 stability Liu et al.
Beclin1 is not the only autophagy protein that is targeted by the UPS in a controlled manner. G-protein-coupled receptor GPCR ligands and agonists were reported to regulate cellular Atg14L levels, and therefore autophagy, through ZBTBmediated ubiquitylation of the protein Zhang T.
et al. Serum starvation increased GSK3β-mediated phosphorylation of ZBTB16, leading to its degradation. Under these conditions, stabilization of Atg14L restored of autophagy.
AMBRA1 is another UPS-controlled autophagy protein. Cullin-4 was identified as an E3 ligase that was responsible for the ubiquitylation of AMBRA1, dooming it for degradation under nutrient-rich conditions where autophagy should be inhibited Antonioli et al.
The PI3K complex subunit p85b is another example. Ubiquitylation of this autophagy signaling component by the E3 ligase SKP1 led to a decrease in its cellular levels and stimulated autophagic activity Kuchay et al. Ubiquitylation of some autophagy proteins did not result in their immediate proteasomal degradation, yet the post-translational modification provided an extra layer of control for the autophagy pathway.
For instance, autophagy receptor OPTN was ubiquitylated as a target of the E3 ligase HACE1, and Klinked ubiquitylation regulated the interaction of the protein with p62 Liu Z. TRAF6, a central E3 ligase of the NF-κB pathway, participated controlled ULK1 activity through Klinked ubiquitylation.
Under nutrient-rich conditions, mTOR phosphorylated AMBRA1 leading to its inactivation. When nutrients were limiting, mTOR inhibition resulted in AMBRA1 dephosphorylation and increased the interaction of the protein with TRAF6.
This event facilitated ULK1 ubiquitylation by TRAF6 Nazio et al. Ubiquitylation of ULK1 resulted in the stabilization of the protein, controlled its dimerization and regulated its kinase activity.
Another ubiquitin-dependent regulation mechanism involved AMBRA1-Cullin-5 interaction in the regulation of mTOR complex component DEPTOR Antonioli et al.
Above-mentioned AMBRA1-Cullin-4 complex dissociated under autophagy-inducing conditions, allowing AMBRA1 to bind another E3 ligase, Cullin This newly formed complex was shown to stabilize DEPTOR and induce mTOR inactivation, providing a negative feed-back loop in the control of autophagy Antonioli et al.
In another study, TLR4 signaling triggered autophagy through Beclin1 ubiquitylation and stabilization. TLR4-associated TRAF6 protein was identified as the E3 ligase responsible for Klinked ubiquitylation of Beclin1 at its BH3 domain.
This modification blocked inhibitory BCL-2 binding to the protein, and free Beclin1 could activate autophagy Shi and Kehrl, On the other hand, the deubiquitinating enzyme A20 reversed TRAF6-mediated ubiquitylation of Beclin1, resulting in autophagy inhibition Shi and Kehrl, Another Klinked ubiquitylation event on Beclin1 was promoted by AMBRA1 protein.
In the same context, the WASH protein interacted with Beclin1, blocked AMBRA1-mediated Beclin1 ubiquitylation, and suppressed autophagy Xia et al.
LC3 and p62 were also subjected to regulatory ubiquitylation. NEDD4 was identified as the E3 ligase in these reactions. NEDD4 was reported to interact with LC3 Sun et al. Moreover, NEDD4 deficient cells exhibited aberrant p62 containing inclusions, indicating the defect in aggresome clearance Lin et al.
Hence, NEDD4 is important for the regulation of p62 function and autophagy. Another essential function of autophagy is the clearance of intracellular pathogens. This special form of autophagy, called xenophagy, is a result of a cooperation between the ubiquitylation machinery and the autophagy pathway.
Pathogens such as Streptococcus pyogenes, Mycobacterium tuberculosis, Listeria monocytogenes, and Shigella flexneri were identified as autophagy targets Gutierrez et al. As a form of selective autophagy, xenophagy involves cargo labeling with ubiquitin, followed by the recognition by autophagy receptors Figure 6.
K and Klinked and linear M1-linked ubiquitin chains were shown to mediate recognition of different pathogens by the xenophagy machinery Collins et al. FIGURE 6. Selective degradation of invaders by xenophagy is example of coregulation of the UPS and autophagy. Cellular degradation of invading bacterium was ubiquilated by various E3 ligases and recognized by adaptor proteins for recruitment autophagic membranes around bacterium.
For example, Salmonella enterica serovar Typhimurium was heavily ubiquitylated in mammalian cells, and activation of xenophagy restricted intracellular bacteria numbers Birmingham et al.
Recent studies showed that, bacterial outer membrane-associated and integral membrane proteins were targets of ubiquitylation Fiskin et al. A number of E3 ligases were involved in xenophagy, including Parkin, RNF, ARIH1, HOIP, and LRSAM1 Huett et al.
For example, both K and Klinked ubiquitylation were observed on Mycobacterium, and Parkin was identified as the E3 ligase catalyzing the Klinked ubiquitylation Collins et al.
Moreover endosome-free areas on the intracellular Salmonella Typhimurium contained a directly attached ubiquitin coat, and addition of linear M1-linked ubiquitin chains by the E3 ligase HOIP of the LUBAC on these ubiquitins contributed to the autophagy of the intracellular parasite Noad et al.
Xenophagy receptors that were described to date include p62, OPTN, NDP52, and NBR1 Thurston, ; Zheng et al. The interplay between ubiquitylation and autophagy achieves the important task of keeping host cells pathogen-free and providing an intracellular innate immune defense mechanism against invaders.
In some reports, ubiquitylated bacteria were found to be surrounded by proteasomes as well Perrin et al. Whether in the elimination of invading organisms, the crosstalk between the UPS and autophagy systems goes beyond ubiquitylation, needs further consideration. As discussed below, cellular mechanisms controlling commensal-turned ancient intracellular microorganisms, namely mitochondria, indeed rely on the function of both the UPS and autophagy.
Mitochondria are vital organelles that form an intracellular dynamic network in the cytosol of eukaryotic cells. Through fusion and fission, they are constantly made and destroyed.
Under steady state conditions, mitochondria might be eliminated by basal in a non-selective manner. On the other hand, elimination of damaged, dysfunctional or superfluous mitochondria requires a selective form of autophagy called mitophagy Lemasters, Programmed elimination of mitochondria during development and differentiation e.
Recent studies showed that mitophagy is a biological phenomenon that involves both the UPS and autophagy. In this section, we will discuss mechanisms of mitophagy, and analyze connections between the UPS and autophagy in this context. Depending on the E3 ligase that ubiquitylates proteins on mitochondria, mitophagy can be divided into two major forms: Parkin-dependent and Parkin-independent mitophagy.
Strikingly, Parkin recruitment to mitochondria was found to be necessary for mitophagy Narendra et al. FIGURE 7. Mitochondrial elimination by autophagy requires the activity of both the UPS and autophagy.
Under normal conditions, after being synthesized as precursor in the cytoplasm, PINK1 was imported to mitochondria by its N-terminal mitochondria targeting sequence MTS. Then, PINK1 was post-translationally modified within mitochondria by resident proteases: MPP and PARL Jin et al.
Cleavage by PARL resulted in destabilization of the protein and its degradation by cytoplasmic proteasomes Yamano and Youle, Under mitochondrial stress however, PINK1 cleavage did not occur and the protein accumulated on the outer mitochondrial membrane OMM Lazarou et al.
Recruitment of cytoplasmic E3 ligase Parkin onto mitochondria required stabilization and the kinase activity of the PINK1 protein Lazarou et al. Parkin itself was a substrate of PINK1 Kondapalli et al. Phosphorylation of Parkin by PINK1 resulted in a conformational change overcoming an autoinhibition, and stimulated its E3 ligase activity Kondapalli et al.
Interestingly, PINK1 was shown to phosphorylate ubiquitin molecules on mitochondrial resident proteins as well. Ubiquitin phosphorylation correlated with an increase in the amount of mitochondria-localized Parkin, providing a feed-forward mechanism of Parkin recruitment Kane et al.
Several proteins on the mitochondrial outer membrane were identified as Parkin ubiquitylation substrates. The list includes VDAC, TOM proteins, mitofusins etc Sarraf et al. Following ubiquitylation some of these targets were shown to be degraded by the proteasome e.
Degradation of proteins related to mitochondrial integrity promoted fission events that facilitate engulfment of mitochondrial portions by autophagosomes, whereas proteins that are not degraded upon ubiquitylation rather contributed to mitochondrial rearrangements e.
The UPS activity was a prerequisite in the preparation of mitochondria for autophagy. Ubiquitylation of mitochondrial targets preceeded the recruitment of the autophagic machinery onto mitochondria Yoshii et al. Serial knock out of putative autophagy receptors showed that NDP52, optineurin OPTN and TAX1BP1 were functional mitophagy receptors, and a triple knockout of these proteins completely blocked mitophagy Lazarou et al.
On the other hand, the autophagy receptor p62 was essential for clustering of damaged mitochondria in perinuclear region of the cells, but not for mitophagy Narendra et al. Ubiquitin modifications on mitochondria might be reversed by the action of DUB proteins.
Several DUBs were identified as positive or negative regulators of mitophagy Dikic and Bremm, ; Wang et al. For example, deubiquitylation of mitochondrial targets by USP15, USP30, and USP35 prevented further progression of mitophagy in a number of cell lines and experimental models Bingol et al.
DUB-mediated deubiquitylation of targets decreased Parkin recruitment onto mitochondria as well Bingol et al. USP8-mediated removal of K6-linked ubiquitin chains from Parkin itself affected recruitment of the protein onto mitochondria and therefore mitophagy Durcan et al. Expression of Parkin is restricted to a few cell types, including dopaminergic neurons.
Consequently, Parkin-null animals showed prominent mitophagy defects only in selected brain regions Lee et al. Therefore in other cell types and tissues, mitophagy has to proceed in a Parkin-independent manner. Alternative E3 ligases were found to play a role in mitophagy in these contexts.
Mulan MUL1 is an E3 ubiquitin ligase that resided on the OMM, and it was shown to play a role in Parkin-independent mitophagy in different model organisms, including Caenorhabditis elegans , Drosophila and mammals Ambivero et al. Mulan stabilized DRP1, led to degradation of MFN2, and interacted with ATG8 family member protein GABARAP Braschi et al.
Another E3 ligase that was associated with mitophagy was GP78 Christianson et al. Over expression of GP78 induced MFN1 and 2 ubiquitylation and degradation, that was followed by mitochondrial fragmentation and mitophagy in cells lacking Parkin Fu et al.
Synphilindependent recruitment of the E3 ligase Siah1 to mitochondria resulted in mitochondrial protein ubiquitylation and mitophagy in a PINK1-dependent but Parkin-independent manner Szargel et al.
Conversely, another OMM E3 ligase, MITOL MARCH5 , was reported to ubiquitylate FIS1, DRP1 Yonashiro et al. All these findings underline the fact that mitophagy might proceed in cells which do not express Parkin. Further studies are required to unravel the molecular mechanisms of Parkin-independent mitophagy in different tissues and cell types, and reveal the details of the crosstalk between the UPS and autophagy under these conditions.
During differentiation, in order to increase their capacity to load hemoglobin-bound oxygen, reticulocytes lose their organelles, including mitochondria, and become mature red blood cells Dzierzak and Philipsen, During this process, a protein called NIX also known as BNIP3L is upregulated Aerbajinai et al.
NIX is a C-terminally anchored outer mitochondrial membrane OMM protein that contains a LC3-interacting region LIR at its cytoplasmic N-terminal part.
Through its LIR domain, NIX interacted with LC3, enabling engulfment of mitochondria by autophagosomes in reticulocytes Novak et al. Characterization of NIX-deficient mice showed that, NIX-deficient Erythrocytes failed to eliminate their mitochondria revealing a critical role for NIX in mitophagy Schweers et al.
Although NIX-dependent mitophagy was predominantly studied in reticulocytes, NIX-dependent mitophagy might be important for other cell types as well [for example, see Esteban-Martínez et al. Autophagy of peroxisomes, pexophagy, is a selective degradation process of peroxisomes during which the UPS and autophagy mechanisms work in collaboration.
Peroxisomes are responsible of a number of cellular functions, including fatty acid oxidation, purine metabolism and phospholipid synthesis Wanders et al.
Several peroxisomal enzymes are involved in redox regulation due to their dual functions in the generation and scavenging of reactive oxygen and nitrogen species. Therefore, peroxisome biogenesis and degradation must be tightly regulated in order to control peroxisome size, number and function Du et al.
Moreover under stress conditions such as hypoxia, oxidative stress, starvation or conditions causing UPS defects, pexophagy is upregulated. During pexophagy, a number of peroxisomal membrane proteins, including peroxins and PMP70 become ubiquitylated Kim et al. PEX2-PEXPEX12 complex serves as an E3 ligase at least for two well studied peroxisome proteins, PEX5 and PMP For example, PEX2 overexpression or amino acid starvation activated the ubiquitylation of PEX5, and another peroxisomal membrane protein, PMP70, and led to peroxisome degradation Sargent et al.
Moreover in response to oxidative stress, ATM was recruited onto peroxisomes through physical interaction with PEX5 and promote its ubiquitylation.
Inactivation of mTORC1 in a TSC2-dependent manner and stimulation of ULK1 phosphorylation by ATM, potentiated pexophagy Zhang J. On the other hand, AAA ATPase complex PEX1, PEX6, and PEX26 was shown to extract ubiquitylated PEX5 from peroxisomal membranes and regulate pexophagy Carvalho et al.
Both NBR1 and p62 were shown to be recruited onto peroxisomes during pexophagy. Yet, NBR1 was a major pexophagy receptor in a number of contexts, and p62 increased the efficiency of NBR1-dependent pexophagy through direct interaction with the latter Deosaran et al.
Altogether, these findings underline the importance of ubiquitylation for the selective degradation of peroxisomes by autophagy.
FIGURE 8. Selective removal of peroxisomes by autophagy utilizes ubiquitylation as signal. In addition to major cellular organelles, autophagy was implicated in the clearance of ribosomes.
Although ribosomes can be degraded in a non-specific manner during non-selective autophagy, a special form of selective autophagy is activated under various stress conditions, and the process is called ribosomal autophagy or ribophagy.
On the other hand, mRNA protein complexes that are stalled during translation form stress granules, and their clearance requires both the UPS and autophagy. In the mammalian system, in addition to mTOR inhibition, oxidative stress, induction of chromosomal mis-segregation, translation inhibition and stress granule formation were all shown to induce ribophagy An and Harper, Ubiquitylation of ribosomes was observed under ER stress-inducing conditions Higgins et al.
Yet, individual ribosomal proteins were indeed shown to be a target of the UPS Wyant et al. NUFIP1-ZNHIT3 proteins were identified as novel ribophagy receptors that directly connected ribosomes to LC3 and autophagy, yet whether ubiquitylation is a prerequisite for ribophagy needs to be clarified by future studies Wyant et al.
FIGURE 9. Ubiquitylation primes ribosomes and stress granules for proteasomal degradation and autophagic elimination. Stress granules are composed of actively accumulated non-translating mRNA ribonucleoprotein complexes Protter and Parker, Proteins that accumulated in the stress granules, include stalled 40S ribosomal units and various translation initiation factors [e.
G3BP1 and TIA-1 are also among the proteins that contribute to stress granule formation Kedersha et al. Moreover, an interplay between G3BP1 and Caprin1 proteins and the DUB protein USP10 was shown to regulate stress granule formation Kedersha et al.
HDAC6 protein was a component of stress granules as well Seguin et al. Endoplasmic reticulum ER stress is one of the conditions under which both the UPS and autophagy pathways are being activated.
Abnormalities in calcium homeostasis, oxidative stress and conditions leading to protein glycosylation or folding defects etc. ER stress might be very destructive for cells, therefore ER-specific stress response pathways such as the unfolded protein response UPR and the ER-associated degradation ERAD pathways were evolved.
Both pathways are directly or indirectly connected to the UPS and autophagy. In mammalian cells, accumulation of unfolded proteins in the lumen of the ER result in the activation of stress responses. PERK activation leads to the phosphorylation of the α subunit of the translation initiation factor, eIF2α, which inhibits the assembly of the 80S ribosome and cap-dependent protein synthesis, while allowing cap-independent translation of the stress response genes such as ATF4.
Activation of IRE1 and ATF6 promotes transcription of other stress response genes. IRE1-mediated processing generates a splice-form of the XBP1 mRNA, resulting in the production of a transcription factor that upregulates chaperones and other relevant genes.
Due to a decrease in the protein load in the ER and an increased folding capacity, the UPR facilitates recovery from stress. In case of failure, the UPR sensitizes cells to programmed death mechanisms.
FIGURE Crosstalk between the UPS and autophagy systems during ER stress and ERAD. Components of the UPR were subject to active regulation by the UPS. For example, SCF component E3 ligase βTrCP was shown to lead to the ubiquitylation ATF4 following its phosphorylation Lassot et al.
CHOP stability was regulated by the UPS and p and cIAP were responsible for CHOP ubiquitylation and degradation counterbalancing its upregulation during ER stress Qi and Xia, ; Jeong et al. Another UPR component, IRE1 was identified as a ubiquitylation target of the E3 ligase CHIP during ER stress.
Ubiquitylation IRE1 inhibited its phosphorylation, perturbed its interaction with TRAF2, and attenuating JNK signaling Zhu et al. Under stress conditions, translation of XIAP, an E3 ligase protein and an inhibitor of apoptosis was downregulated in a PERK-eIF2α-dependent manner.
In the same context, ATF4 may promote ubiquitylation and degradation of XIAP, leading to sensitization of cells to ER stress-related cell death Hiramatsu et al.
Conversely, activation of PERK-eIF2α axis might also show opposing effects through induction of other IAP proteins, cIAP1 and cIAP2, and counter balance cell death induction signals Hamanaka et al.
Endoplasmic reticulum stress was shown to trigger autophagy, and ER-related stress response mechanisms were involved in the process. PERK-mediated phosphorylation of eIF2α and resulting ATF4 and CHOP activation, were associated with the transcription of genes such as ATG5, ATG12, Beclin1, ATG16L1, LC3, p62 and TSC2 activator, hence mTOR inhibitor REDD1 Whitney et al.
Moreover, CHOP downregulated BCL2 binding Mccullough et al. TRB3, an AKT inhibitor protein, was also described as a target of CHOP Ohoka et al. In addition, IRE1 activation resulted in the recruitment of ASK1 by the adaptor TRAF2 and the outcome was the activation of JNK and p38 kinases Nishitoh et al.
BCL2 is one of the targets of JNK, its phosphorylation by the kinase resulted in destabilization the inhibitory BCL2-Beclin1 complex, stimulating autophagy Bassik et al.
On the other hand, in its unspliced form, IRE1 splicing target XBP1, in its unspliced form was shown to target the autophagy activator FOXO1 for degradation by the UPS Vidal et al. Endoplasmic reticulum is a major calcium store in cells, and calcium release to cytosol was observed during ER stress.
In addition to problems with SERCA refill pumps and leakiness of membranes during stress, upregulation of ERO1-α by CHOP resulted in an IP3-mediated calcium release Li et al.
Calcium binding protein calmodulin senses the cytosolic increase in the concentration of the ion, and bind to calmodulin-regulated kinases such as CaMKII and DAPK1, modulating their activity.
Activated CaMKII was shown to stimulate autophagy through AMPK phosphorylation and activation Høyer-Hansen et al. In addition, calmodulin-binding and PP2A-mediated dephosphorylation was necessary for the activation of the autophagy-related kinase DAPK1 Gozuacik et al.
DAPK1 could directly phosphorylate Beclin1 on the BH3-domain, resulting in the dissociation of Beclin1 from the BCL2-Beclin1 complex and allowing it to stimulate autophagy Zalckvar et al.
Proteins that accumulate in the ER are degraded by the ER-associated degradation ERAD system. ERAD mediates transport, extraction and ubiquitylation of proteins that cannot be salvaged and target them for degradation in proteasomes. In mammalian cells, ER membrane-resident complexes containing E3 ligases such as HRD1 and GP78, and other regulatory components such as EDEM1, SEL1L, ERManI, and HERP control the ERAD pathway.
DUB proteins, including YOD1, USP13, USP19, and Ataxin-3 were implicated in the control of client protein ubiquitylation and ERAD substrate modulation Zhong and Pittman, ; Bernardi et al. ER-associated degradation regulators and therefore ERAD might be controlled by the UPS and autophagy pathways.
For example, E3 ligase Smurf1 was found to be downregulated during ER stress, resulting in the accumulation of its direct ubiquitylation target WFS, which is a stabilizer ER-related E3 ligase HRD1 Guo et al.
Smurf1 was also involved in selective bacterial autophagy Franco et al. On the other hand, while the ERAD complex component HERP protein was degraded by the UPS Hori et al. An ER-localized E3 ligase synoviolin protein was shown to ubiquitylate HERP protein and control its degradation by proteasome Maeda et al.
Yet, other ERAD-related components, EDEM1 and Derlin2 as well as ubiquitylated EDEM1 proteins colocalized with cytoplasmic aggregates and autophagy receptors p62 and NBR1, they were degraded by selective autophagy Le Fourn et al. ERManI, a mannosidase that is responsible for priming ER-resident glycosylated proteins for degradation, was described as an accelerator of the ERAD pathway and clearance of clients by the UPS.
But, following proteasome inhibition and subsequent ER stress, ERManI colocalized with LC3 and degraded in an autophagy-dependent manner Benyair et al. All these findings point out to the presence of important junctions and coregulation nodes between the UPS and autophagy in the context of ER stress.
Additionally, ERphagy, the autophagy of portions of the ER, was implicated in the recovery from ER stress and control of ER size, but this mechanism was so far described as a ubiquitin-independent process Schuck et al.
Several transcription factors that are regulated by the UPS, including p53, NFκB, HIF1α, and FOXO, have been implicated in the control of autophagy.
In general, these factors were shown to directly activate transcription of key autophagy genes under stress conditions. Some autophagy proteins such as LC3 are consumed in the lysosome following delivery, and during prolonged stress, cellular levels of these proteins are sustained by mechanisms, including transcription.
On the other hand, regulation of the transcriptional activity NRF2 involves a special crosstalk between the two systems. In this section, we will summarize molecular details of transcription regulation by the UPS and autophagy. P53, a guardian of the genome, is one of the well-known transcriptonal regulators that has a dual role in autophagy depending on its intracellular localization.
Accumulating p53 protein activates transcription of several stress- and death-related genes, including autophagy-related genes PRKAB1 , PRKAB2, TSC2 , ATG2, ATG4, ATG7 , ATG10 , ULK1 , BNIP3, DRAM1, and SESN2 Crighton et al.
On the other hand, a cytosolic form of p53 was shown to inhibit AMPK and activate the mTOR pathway. Additionally, another E3 ligase, NEDD was shown to control MDM2 stability and p53 activation Xu et al. In addition to MDM2, another E3 ligase, PIRH2, was able to ubiquitylate p53 to control its cellular stability Shloush et al.
NF-κB is a well studied transcriptional regulator of autophagy. As a result of its association with IκB, NF-κB is found in an inactive state in the cytosol. In response to agonists, IκB was reported to be ubiquitylated and subsequently degraded by the UPS.
Phosphorylated IκB recruits the E3 ligase SCF-βTRCP, followed by its degradation in the proteasome Orian et al. After IκB degradation, NF-κB was then free to migrate to the nucleus of the cell, and induce transcription of target genes, including Beclin1 and p62, and induce autophagy Copetti et al.
Another level of regulation involved TNF-α receptor-associated protein complexes. Ubiquitylated RIPK1 could recruit NEMO and TAB-TAK1 complex for IKK activation and hence NF-κB stimulation. Additionally, RIPK1 could also be modified by A20 through addition of Klinked poly-ubiquitin chains, sending the kinase for proteasomal degradation Kravtsova-ivantsiv et al.
However, in some contexts, TNF-α-induced NF-κB activation was reported to inhibit autophagy Djavaheri-Mergny et al. Furthermore in some contexts, RIPK1 silencing activated autophagy under both basal and stress conditions Yonekawa et al.
On the other hand, RIPK1 itself was reported to be a target of pmediated selective autophagy Goodall et al. Moreover, autophagy was responsible for the degradation of NF-κB activator NIK and IKK complex subunits, indicating the presence of a tight cross-regulation of the NF-κB pathway by the UPS and autophagy Qing et al.
Another transcription factor that was controlling the autophagic outcome was HIF1α. Oncogene ; 19 — Stabilization of basally translated NF-kappaB-inducing kinase NIK protein functions as a molecular switch of processing of NF-kappaB2 p Qu Z, Qing G, Rabson A, Xiao G.
Tax deregulation of NF-kappaB2 p processing involves both beta-TrCP-dependent and -independent mechanisms. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p Mol Cell ; 8 — Xiao G, Civijic ME, Fong A, et al.
EMBO J ; 20 — Qing G, Xiao G. Essential role of IkappaB kinase alpha in the constitutive processing of NF-kappaB2 p Basso AD, Solit DB, Chiosis G, et al. Akt forms an intracellular complex with heat shock protein 90 Hsp90 and Cdc37 and is destabilized by inhibitors of Hsp90 function.
Broemer M, Krappmann D, Scheidereit C. Requirement of Hsp90 activity for IkappaB kinase IKK biosynthesis and for constitutive and inducible IKK and NF-kappaB activation. Oncogene ; 23 — Chowdary DR, Dermody JJ, Jha KK, Ozer HL. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway.
Mol Cell Biol ; 14 — Orlowski M, Wilk S. Ubiquitin-independent proteolytic functions of the proteasome. Arch Biochem Biophys ; :1—5. Edinger AL, Thompson CB.
Defective autophagy leads to cancer. Cancer Cell ; 4 — Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ ; 12 Suppl — Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword.
Science ; — Samari HR, Seglen PO. Inhibition of hepatocytic autophagy by adenosine, aminoimidazolecarboxamide riboside, and N6-mercaptopurine riboside.
Evidence for involvement of amp-activated protein kinase. Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period.
Yu L, Wan F, Dutta S, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA ; — Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation.
J Cell Sci ; — CAS PubMed Google Scholar. Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy.
Nat Rev Cancer ; 5 — Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest ; — Download references. We thank Drs Covey L for the Ramos cells, Mizushima N for Atg5 null MEFs and Ozer HL for ts20 cells.
This study was assisted partially by research grants from the New Jersey Commission on Cancer Research, Fifth District AHEPA Cancer Research Foundation, American Cancer Society RSG MGO and National Institutions of Health 1R01 CA to GX.
Department of Cell Biology and Neuroscience, Rutgers, The State University of New Jersey, Allison Road, Piscataway, , NJ, USA. You can also search for this author in PubMed Google Scholar.
Correspondence to Gutian Xiao. Reprints and permissions. Qing, G. Hsp90 inhibition results in autophagy-mediated proteasome-independent degradation of IκB kinase IKK.
Cell Res 16 , — Download citation. Received : 28 September Revised : 09 October Accepted : 10 October Published : 07 November Issue Date : 01 November Anyone you share the following link with will be able to read this content:.
Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative. Cellular and Molecular Life Sciences Skip to main content Thank you for visiting nature.
nature cell research original article article. Download PDF. Abstract Autophagic and proteasomal proteolysis are two major pathways for degradation of cellular constituents. Identification of a phosphorylation site on Ulk1 required for genotoxic stress-induced alternative autophagy Article Open access 09 April A novel ATG5 interaction with Ku70 potentiates DNA repair upon genotoxic stress Article Open access 17 May Introduction Cellular homeostasis requires a well-controlled balance between protein synthesis and degradation.
Materials and Methods Expression vectors and reagents Expression vectors encoding IKK have been described as before Cell culture and transfection Human B-cell line Ramos RG69, mouse fibroblasts ts20 and Atg5 knockout mouse embryonic fibroblasts MEFs were gifts from Drs Covey L, Ozer HL and Mizushima N, respectively.
Immunoblotting Cells were lysed in radioimmuoprecipitation assay buffer RIPA buffer 50 mM Tris-HCl pH 7. Results Hsp90 is required for protein expression of IKK Although it is clear that Hsp90 physically associates with IKK, the role of Hsp90 in IKK expression is still controversial 7 , Figure 1.
Full size image. Figure 2. Figure 3. Figure 4. Discussion The proteasome and autophagy are two highly conserved mechanisms that are primarily employed for protein degradation within eukaryotes.
Figure 5. Similar content being viewed by others. Abbreviations AICAR: 5-aminoimidazolecarboxamide 1-b-D-ibofuranoside CMA: chaperone-mediated autophagy GA: geldanamycin Hsc heat-shock cognate of 70 kDa Hsp heat-shock protein of 90 kDa IκB: inhibitor of NF-κB IKK: IκB kinase.
References Levine B, Klionsky DJ. Article CAS Google Scholar Lodish H, Berk A, Matsudaira P, et al. Google Scholar Dai C, Whitesell L. Article CAS Google Scholar Zhang H, Burrows F. CAS Google Scholar Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM.
Metrics details. Autophagy and proteasomal degradation ubiquitin—proteasome system UPS and autophagy are Aktophagy distinct Autophagy and proteasomal degradation interacting proteolytic Autohagy. They play critical roles Anxiety self-help tips cell survival under normal conditions and during stress. An increasing body of evidence indicates that ubiquitinated cargoes are important markers of degradation. p62, a classical receptor of autophagy, is a multifunctional protein located throughout the cell and involved in many signal transduction pathways, including the Keap1—Nrf2 pathway. Autophagy and the ubiquitin—proteasome system UPS are the proteaso,al major Immune system protection supplements quality control and recycling Proteasomwl that are responsible Autophagy and proteasomal degradation cellular homeostasis in eukaryotes. Ubiquitylation is utilized as a degradation signal by Autophagy and proteasomal degradation systems, yet, pgoteasomal mechanisms are in xnd. The UPS anr responsible for the degradation of short-lived proteins and soluble misfolded proteins whereas autophagy eliminates long-lived proteins, insoluble protein aggregates and even whole organelles e. Both the UPS and selective autophagy recognize their targets through their ubiquitin tags. In addition to an indirect connection between the two systems through ubiquitylated proteins, recent data indicate the presence of connections and reciprocal regulation mechanisms between these degradation pathways. In this review, we summarize these direct and indirect interactions and crosstalks between autophagy and the UPS, and their implications for cellular stress responses and homeostasis.
0 thoughts on “Autophagy and proteasomal degradation”